Importing custom data formats
Sergio Oller
2025-05-03
importing-custom-data-formats.RmdAbstract
This vignette shows how to import your custom data so it can be used with the GCIMS package. Data formats are typically vendor-dependant, and exports to CSV can have subtle differences.
Introduction
This vignette aims to show you how to create a GCIMSDataset object from your own files, if those are not supported natively by the GCIMS package.
We do so, by showing how we can add support for importing CSV files.
The first step is to read the drift time, the retention time and the intensity matrices from your data file. Then we create a GCIMSSample object.
Once we have solved that, we wrap all our written code into a function, and we create the dataset.
Creating GCIMSSample objects
To create a GCIMSSample object you need to have at least:
- A numeric vector with drift times in ms
- A numeric vector with retention times in s
- An intensity matrix, with dimensions
If your time vectors have different units, GCIMS will work, although you may see wrong labels in plots. We plan to include support for more units in the future.
Let’s imagine your sample is on a CSV file, with retention times on the first column, drift times on the first row, and the corresponding intensity values.
We will now create two samples: sample1.csv and sample2.csv
your_csv_file <- (
",0.0,0.1,0.2,0.3,0.4
0.0, 0, 20, 80, 84, 23
0.8,123,200,190,295, 17
1.6,230,300,200, 92, 15
2.4,120,150,120, 33, 22
3.2, 70,121, 74, 31, 34
")
write(your_csv_file, "sample1.csv")
write(your_csv_file, "sample2.csv")You can read it using read.csv() or the
readr::read_csv() function from the readr
package.
your_csv_file <- "sample1.csv"
csv_data <- read.csv(your_csv_file, check.names = FALSE)Once loaded, your data will look like:
csv_data
0.0 0.1 0.2 0.3 0.4
1 0.0 0 20 80 84 23
2 0.8 123 200 190 295 17
3 1.6 230 300 200 92 15
4 2.4 120 150 120 33 22
5 3.2 70 121 74 31 34- The first column contains the retention time.
- The column names (with the exception of the first column, which is empty) contain the drift time
- The values of all columns but the first are the intensities
retention_time <- csv_data[[1]]
drift_time <- as.numeric(colnames(csv_data)[-1])
intensity <- as.matrix(csv_data[,-1])
rownames(intensity) <- retention_timeThe retention time:
The drift time:
The intensity matrix:
intensity
0.0 0.1 0.2 0.3 0.4
0 0 20 80 84 23
0.8 123 200 190 295 17
1.6 230 300 200 92 15
2.4 120 150 120 33 22
3.2 70 121 74 31 34With these three elements, we can create a GCIMSSample:
s1 <- GCIMSSample(
drift_time = drift_time,
retention_time = retention_time,
data = intensity
)
s1
A GCIMS Sample
with drift time from 0 to 0.4 ms (step: 0.1 ms, points: 5)
with retention time from 0 to 3.2 s (step: 0.8 s, points: 5)We are now ready to define a parser function that
returns a GCIMSSample given a filename:
GCIMSSample_from_csv <- function(filename) {
csv_data <- read.csv(your_csv_file, check.names = FALSE)
retention_time <- csv_data[[1]]
drift_time <- as.numeric(colnames(csv_data)[-1])
intensity <- as.matrix(csv_data[,-1])
rownames(intensity) <- retention_time
return(
GCIMSSample(
drift_time = drift_time,
retention_time = retention_time,
data = intensity
)
)
}Try it with a single sample:
s1 <- GCIMSSample_from_csv("sample1.csv")
s1
A GCIMS Sample
with drift time from 0 to 0.4 ms (step: 0.1 ms, points: 5)
with retention time from 0 to 3.2 s (step: 0.8 s, points: 5)You can check the intensity matrix and you can plot the sample to check that it behaves as expected:
intensity(s1)
rt_s
dt_ms 0 0.8 1.6 2.4 3.2
0 0 20 80 84 23
0.1 123 200 190 295 17
0.2 230 300 200 92 15
0.3 120 150 120 33 22
0.4 70 121 74 31 34
plot(s1)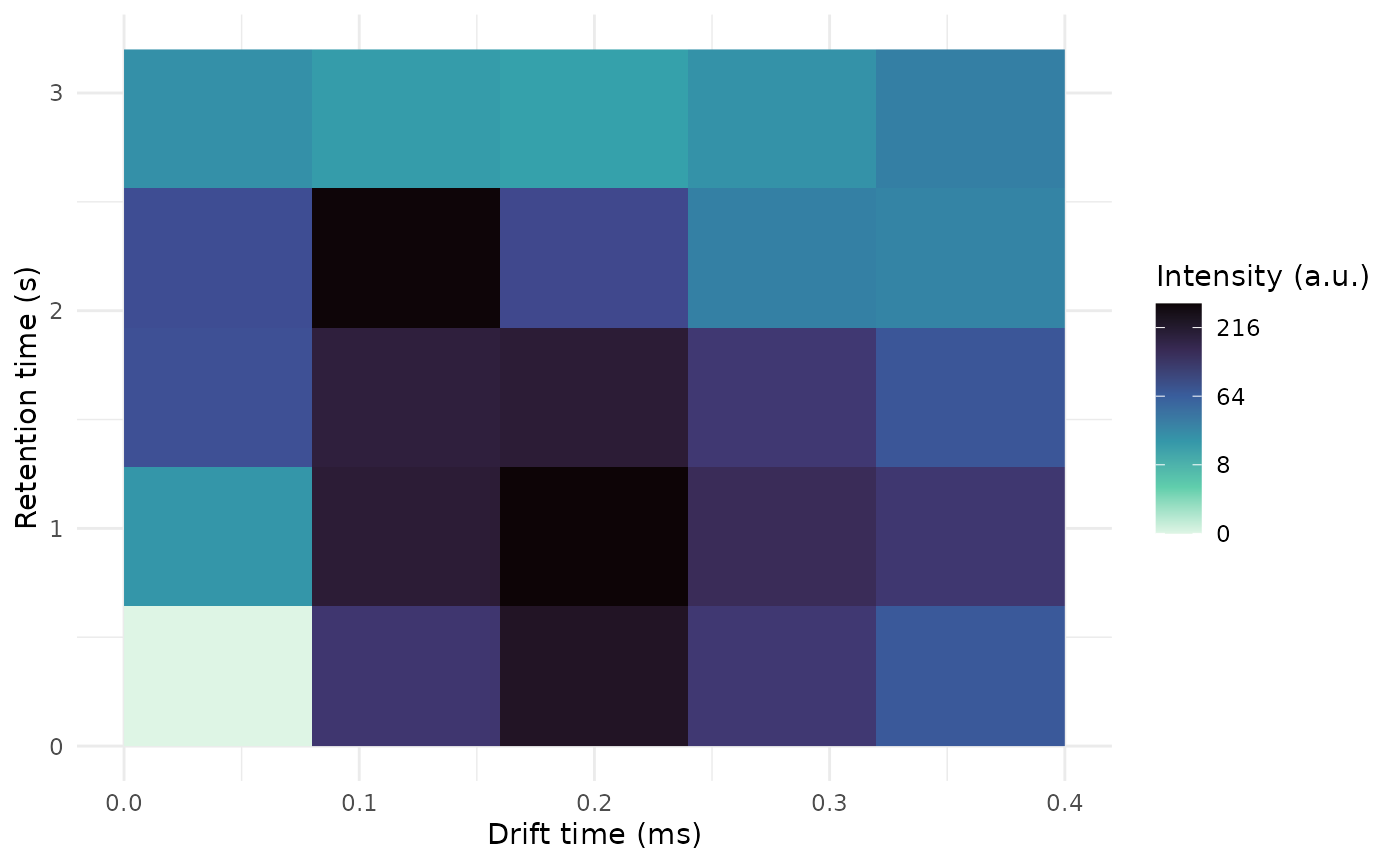
Create the GCIMSDataset
Once you are satisfied with your function, prepare the phenotype data frame:
pdata <- data.frame(
SampleID = c("Sample1", "Sample2"),
FileName = c("sample1.csv", "sample2.csv"),
Sex = c("female", "male")
)
pdata
SampleID FileName Sex
1 Sample1 sample1.csv female
2 Sample2 sample2.csv maleAnd create the dataset object, passing your parser
function:
ds <- GCIMSDataset$new(
pData = pdata,
base_dir = ".",
parser = GCIMSSample_from_csv,
scratch_dir = "GCIMSDataset_demo1"
)
ds
A GCIMSDataset:
- With 2 samples
- Stored on disk (not loaded yet)
- No phenotypes
- No previous history
- Queued operations:
- read_sample:
base_dir: /__w/GCIMS/GCIMS/vignettes
parser: < function >
- setSampleNamesAsDescription
You now have a dataset ready to be used.
Session info
sessionInfo()
R version 4.4.1 (2024-06-14)
Platform: x86_64-pc-linux-gnu
Running under: Ubuntu 22.04.4 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
time zone: UTC
tzcode source: system (glibc)
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] cowplot_1.1.3 GCIMS_0.1.1 BiocStyle_2.32.1
loaded via a namespace (and not attached):
[1] sass_0.4.10 generics_0.1.3 digest_0.6.37
[4] magrittr_2.0.3 evaluate_1.0.3 grid_4.4.1
[7] RColorBrewer_1.1-3 bookdown_0.43 fastmap_1.2.0
[10] jsonlite_2.0.0 ProtGenerics_1.36.0 BiocManager_1.30.25
[13] purrr_1.0.4 viridisLite_0.4.2 scales_1.4.0
[16] codetools_0.2-20 textshaping_1.0.1 jquerylib_0.1.4
[19] cli_3.6.5 rlang_1.1.6 Biobase_2.64.0
[22] withr_3.0.2 cachem_1.1.0 yaml_2.3.10
[25] parallel_4.4.1 tools_4.4.1 BiocParallel_1.38.0
[28] dplyr_1.1.4 ggplot2_3.5.2 sgolay_1.0.3
[31] BiocGenerics_0.50.0 vctrs_0.6.5 R6_2.6.1
[34] stats4_4.4.1 lifecycle_1.0.4 S4Vectors_0.42.1
[37] fs_1.6.6 htmlwidgets_1.6.4 MASS_7.3-65
[40] ragg_1.4.0 pkgconfig_2.0.3 desc_1.4.3
[43] pkgdown_2.1.2 bslib_0.9.0 pillar_1.10.2
[46] gtable_0.3.6 glue_1.8.0 systemfonts_1.2.3
[49] xfun_0.52 tibble_3.2.1 tidyselect_1.2.1
[52] knitr_1.50 farver_2.1.2 htmltools_0.5.8.1
[55] rmarkdown_2.29 labeling_0.4.3 signal_1.8-1
[58] compiler_4.4.1